|
Postdoc Kari Goodman's paper on Nesosydne divergence is out in Evolution. Here's the link. Congratulations!! And happy National Postdoctoral Appreciation Week!
0 Comments
Check out Jessica's paper reviewing a new book on Phylogenetic Networks in the journal Taxon. Congratulations!
 Departmental coffee hours are supposed to be cordial affairs, a chance to catch up with colleagues and exchange news and ideas. So why was this graduate student I’d just met getting up in my face about using the “wrong” locus for my work characterizing fungal communities in Hawaii? I’ve been using the D1/D2 domain of 26S rRNA and finding an amazing amount of diversity in plant substrates Hawaiian drosophilids use for reproduction, oviposition, and larval development. The grad student seemed to think if I wasn’t pyrosequencing using ITS, I was just wasting my time. But I’m not pyrosequencing, although I am accepting contributions from benevolent patrons to do so. Instead, I’m focusing on one locus that will be informative over wide phylogenetic space without becoming saturated with mutations. In a recent paper comparing the utility of D1/D2 and ITS for yeast species delimitation, Groenewald et al (2011) found 3 to 4 times greater nucleotide diversity in ITS. That worked out great for their work, in which they combined the sequences from both loci with morphology, mating studies, and physiology to propose three new yeast species in the genus Candida. In another study, Zimmerman and Vitousek used only ITS to identify over 4200 fungal OTUs across 13 sites on Mauna Loa. BLAST searches showed these OTUs spread over a dozen or so fungal classes, but most were either Dothideomycetes (36%) or Not assigned/environmental sample (40%). The problem I see with this is that they identified 4200+ OTUs BLASTing to distant classes using 157 bp of sequence. The experiments were rigorously carried out, but saturation of variable sites, where the same base location is hit with the same mutation in multiple lineages, must be an issue. That can cause errors in defining OTUs, making less related taxa appear to be more related. So in fact Zimmerman and Vitousek may have picked up less diversity than was actually present in their samples. 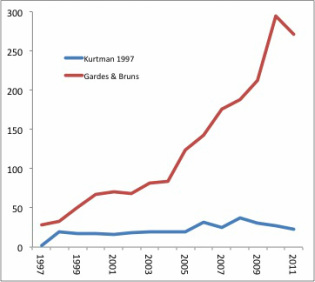 Getting back to the grad student’s criticism, is the D1/D2 domain obsolete? I did a quick search on Web of Science to find out. I searched for citations of the paper in which the NL1 and NL4 primers, which amplify the domain, were first described (Kurtzman 1997), and on the paper (Gardes & Bruns 1993) in which one of the primers, ITS1-F, used by Zimmerman & Vitousek (2012) was presented. These searches gave the following hits, broken down by year. Kurtman’s D1/D2 domain primers have seen pretty steady use over the last decade, with a slight uptick in the late 2000’s. Use of the ITS primer has steadily increased over the same period. Why? I think it’s a combination of one locus becoming accepted as the “standard”, and the fact that there is more phylogenetic information (variation) contained within the ITS locus, which is probably why it became the standard. In addition, the sequence length works quite well with high-throughput pyrosequencing, allowing the generation of very large data sets, as in Zimmerman & Vitousek (2012). So, which locus would you use? I still think it makes sense to choose a less variable locus when you’re searching across wide phylogenetic space. But to make more refined identifications to the level of species, I’d use ITS, and I’d like to make the switch to pyrosequencing to do it. Brian Ort The study of how people interact with their environments is a fascinating field in which the interplay of traditional ecological knowledge, environmental psychology, and justice are explored. Although ethnobotany is relatively well-covered ground in scientific literature, ethnomycology still quite unchartered territory. Scholarly articles on the traditional uses of fungi as food, medicine, and ceremonial usage of hallucinogenic mushrooms have focused mostly on indigenous tribes in developing nations. Fascinating accounts have been made on the usage of psychoactive Psilocybe in Thailand (Gartz, 1994), the collection of on average the 8 species of edible mushrooms by Igbo women and children in Nigeria (Akpaja,, 2003), and the mysticisms associated with the infamous Amanita muscaria in a number of Latin America, Hindu, and even Roman culture (Lowy, 1974). Considering that fungi have be used in a myriad of ways, such as fermenting alcohol, in recipes, as medicines, in many countries for such a long time, it is interesting that some cultures literally fear mushrooms. ‘Fungiphobia’ is a fascinating phenomenon that primarily plagues the United States and Great Britain. The fear of fungi primarily stems from the caution against consuming poisonous wild mushrooms, such as the Fly Agaraics. But upon further investigation we find that the death toll associated with mushroom poisoning is extremely low. In North American in 2009, there were 8 deaths caused by mushroom poisoning, 4 of those deaths were recorded in dogs (NAMA). Furthermore, of the 10,000 described species of mushrooms, roughly 400 species are poisonous, and of those, only 20 are common! Thus the consumption of poisonous mushrooms could be easily avoided with increased knowledge of mushroom identification. A number of European countries like Italy and France are quite ‘fungiphillic’ and appreciates a wide diversity of wild collected mushrooms in cooking. In the U.S. commercial production of mushrooms utilizes very few species. In fact, the three most highly produced mushrooms are all varieties of the same species, Agaricus bisporus, the button mushroom, AKA portabella and crimini. It is clear that a number of bioculutral services could be gained by setting aside our fungiphobia and delving embracing ethnomycology, if only to spice up our meals.
Iman Sylvain How do new species form? This pervasive question in evolutionary biology has its roots in Darwin (and let’s not forget the common man’s evolutionist – Wallace) and natural history observation, but has flourished over the last couple of decades with modern molecular and computational techniques. Birds, fish, lizards and arthropods get most of the attention on this subject, while an incredibly diverse member of the eukaryotes, the fungi, has received very little.
How diverse are the fungi? Educated guesses are variable, and currently range between 1.5 million and 5.1 million. With all that diversity, and so many examples of fungi that intersect with our lives (for some great stories on this, see the Cornell Mushroom Blog) it is incredible how little attention they have received as the subjects of speciation studies. Why? I could chalk it up to the charismatic macrofauna effect, but c’mon – there is a fungus that turns ants into zombies! And then there’s another fungus that prevents that zombie fungus from spreading out of control! The world is full of charismatic fungi. Alternatively, the reason is that as humans we can more or less picture what an animal species is while this is not necessarily so with the fungi - we typically can’t even picture an entire fungal individual. Among folks who study how species form, there has been a particular lot of attention focused on the role that adaptation to different resources plays in the fracturing of species. It is now being suggested that fungal plant pathogens are incredible model systems for this area of research, as one of the primary ways new plant fungal pathogen species may emerge is by shifting onto new host plant resources. This is interesting for at least two reasons. First, the large amount of theory and research that exists on speciation may be able to help guide plant pathogen researchers at understanding the internal factors controlling host switches in the fungal pathogens. Second, it points speciation researchers towards a myriad of new study systems on which experiments can be done. Tatiana Giraud and colleagues point out in a 2010 article that plant pathogens are full of characteristics that make them great for studying rapid ecological speciation (their hosts provide strong selection, they have lots of offspring which increases both their survival and mutation rate, they mate entirely within the host, they have only a few genes that control host specificity and they can clone themselves to keep themselves going for long periods between mating with another individual). How rapid is rapid? In one example, a rust fungal species emerged by hybridization IN 1997 to infect a poplar tree that itself is a hybrid (Newcombe et al 2000). Wow! In another recent example, researchers studied a pathogen that infects green coffee berries, and found that it diverged approximately 2200 yrs ago. Although ages longer than the first example, this is still very recent in the realm of speciation examples, and leads me to wonder what else is going on out there in this fast-paced world of fungal speciation. From the perspective of an insect researcher, where experiments can be done but lab rearing is tricky, this is fascinating stuff. Stay tuned, and especially to the Giraud Lab (Ecologie, Systematique et Evolution, Universite Paris-Sud) for more neat work in this area! Kari Goodman Newcombe, G, Stirling, B., McDonald, S., and Bradshaw, H.D., Jr. (2000) Melampsora x columbiana, a natural hybrid of M. medusae and M. occidentalis. Mycological Research 104(3): 261-274.  Yeasts and other fungi are important components of decomposer commnunities and may have large impacts on rates of diversification in saprophagous insect taxa. Two Drosophila systems have been investigated in detail; morning glories in the Hawaiian Islands and the cacti in the southwesten United States. These two systems are the best understood examples of symbiotic relationships among flies, their host plants and the microbes that make the plant tissue suitable for larval development. Herman Phaff began surveying Hawaiian yeasts in collaboration with William B. Heed (Speith 1981). Heed had worked on the ecological associations of Hawaiian Drosophilidae, focusing mainly on rearing flies from various host plants (Heed 1968; 1971). He was interested in delving into more detail on this system as part of his sabbatical at UC Davis. During this time he also worked with Phaff on the cactophilic Drosophila in the southwesten United States. The Phaff Collection at Davis still consists of many of the yeasts discovered as part of this research. Lachance, Starmer and their colleagues (Lachance et al. 1990; 2001; 2003; 2005) picked up where Phaff and Heed left off, describing many yeast species present in Hawaii, as well as their specific interactions with plants and insects. They studied the relatively simple flower-yeast-insect systems using culture dependent approaches. Recently, several studies (Ort, et al. 2012; Zimmerman and Vitousek 2012) have expanded our knowledge of Hawaiian fungal and yeast systems, using culture-independent, DNA sequence-based approaches to examine communities in several native plant genera (e.g., Metrosideros, Clermontia, Pisonia, Acacia, Cheirodendron). Research on Hawaiian fungi are still in the descriptive stage but once these systems are better understood, a wealth hypotheses addressing biogeography, coevolution and adaptive radiation, and community assembly can be tested. Patrick O'Grady Recent advances in sequencing technologies have allowed researchers to describe microbial richness in very fine resolution. Typically sequences are categorized into OTUs (operational taxanomic units) based on 95%, 98%, or 99% similarity between sequences. For example, Zimmerman and Vitousek found 4063 fungal endophyte OTUs (95% cutoff) on the leaves of one species of tree (Metrosideros polymorpha) at 13 sites (10 trees per site). Even with a tremendous number of OTUs, this and other microbial genetic surveys conclude that sampling may be insufficient as is evident by the lack of asymptotic response of rarefaction curves. What is the “usefulness” of this number of OTUs from an ecological perspective, in other words what does this number of species “do” for the system? To this end it might be useful to assign functionality to groups of OTU’s so that functional diversity could be incorporated into these studies. This thought led me to a review of fungal endophytes function by Rodriguez et al. 2009 in the New Phytologist. As a fungal neophyte (not endophyte) I found the review helpful.
Here are a few highlights:
Clearly, before functional diversity is assigned to OTU groups for ecological comparisons, a great deal of work needs to be done to determine the roles of the highly diverse fungal endophyte groups. Lisa Marrack While some insects can survive more than 10x the amount of radiation than humans, thank you MythBusters, the real inheritors of the nuclear apocalyptic world would have to be fungi. Why? Because radiation exposure for insects would continue after their initial exposure by seeping in through their food sources, but for a fungus that radiation would be lunch!
But how is this possible? First, the numbers of fungi inhabiting planet earth is immense. They are everywhere and you’ve probably experienced them in the form of dandruff or the dreaded athlete’s foot. So when black fungus was found growing on the walls of the still highly radioactive Chernobyl reactor, it wasn’t too exciting, until Dr. Auturo Casadevall began to wonder if the fungus was actually using the radiation as an energy/food source. “But I thought fungi were decomposers”, you might be saying to yourself, and you’d be right. Because fungi lack chlorophyll, fungi cannot synthesize their own energy source and therefore must consume organic compounds for fuel. Then how might fungus incorporate radiation, something very akin to sunlight, into their metabolism? The answer is melanin. Yup, I do mean the melanin in our skin that protects us from sunburns (or lack there of for our fair-skinned friends). However, fungi seem to use their melanin for energy capture rather than the acquisition of the perfect summer tan. Studies show that when the melanized fungal cells of Wangiella dermatitidis and Cryptococcus neoformans encounter 500 times the amount of background radiation they actually grow faster even under nutrition-limited conditions unlike their unmelaninized counterparts. So, when and if a nuclear holocaust does occur, the cockroaches will have plenty of food to munch on. In the meantime, science could utilize these melanin-rich fungi to expand our knowledge of energy capture in the human body, or even radiation clean-up, currently an unexplored arena. While I don’t expect fungi to sprout legs, don a spidey suit, and fight nuclear-aftermath crimes, I do think they are a start to recolonizing highly radioactive areas and that seems close enough to a super hero power. Jessica Craft This article looks at the changes in endophyte communities in leaves of a single species of tree in different environments on Mauna Loa Volcano, Hawai’i. According to this article, the phyllosphere, or the surface and interior of leaves, is one of the least studied environments in the world (even though its area is approximately twice that of land surface!). The phyllosphere also happens to host an incredibly high number of fungal endophytes, as discovered by this study. Specifically, this study looked at operational taxonomic units (OTU’s), and determined that there were over 4,200 OTU’s present at the 13 study sites, all of which were found on the same species of tree, and within 80 km of each other. This richness in OTU’s represents roughly half of the number of OTU's found in a global study of indoor fungi-which is a huge amount of diversity! And all of this diversity was found on a single host species. Clearly, there is potential for a much higher level of fungal diversity than previously thought.
Aside from finding a huge amount of fungal diversity, this study also found that environmental variables are a factor in determining the fungal endophyte community of the phyllosphere. The environmental variables that were looked at were rainfall, elevation, and the age of the lava that the site/tree was located on. The study found that fungal endophytic communities were more similar with similar amounts of rainfall and elevation; thus communities correlated with these environmental variables. This finding is important for a number of different applications, including carbon fixation, community ecology, and climate change. Finding out more about how differences in temperature and rainfall will change the phyllosphere and how it functions could help us predict and understand yet another possible effect of climate change. Additionally, looking at the roles of these many different species of fungi in the phyllosphere community could help us design more effective solar sequestration methods. If I wasn’t already betrothed to a PhD project looking at barcoding of benthic macroinvertebrates and biomonitoring of aquatic ecosystems, I would definitely be interested in looking at the fungal endophytic communities within the phyllosphere! Natalie Stauffer NB Zimmerman and PM Vitousek (2012). Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape |
PatrickProfessor Archives
April 2018
Categories
All
|
| O'Grady Lab |
|
 RSS Feed
RSS Feed
