
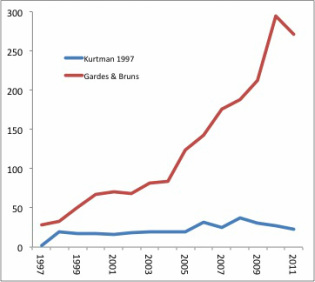
Kurtman’s D1/D2 domain primers have seen pretty steady use over the last decade, with a slight uptick in the late 2000’s. Use of the ITS primer has steadily increased over the same period. Why? I think it’s a combination of one locus becoming accepted as the “standard”, and the fact that there is more phylogenetic information (variation) contained within the ITS locus, which is probably why it became the standard. In addition, the sequence length works quite well with high-throughput pyrosequencing, allowing the generation of very large data sets, as in Zimmerman & Vitousek (2012).
So, which locus would you use? I still think it makes sense to choose a less variable locus when you’re searching across wide phylogenetic space. But to make more refined identifications to the level of species, I’d use ITS, and I’d like to make the switch to pyrosequencing to do it.
Brian Ort
 RSS Feed
RSS Feed
