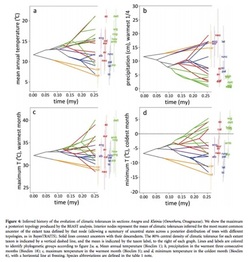
One new ancestral state reconstruction method I recently learned uses the software package Phyloclim maintained by Christoph Heibl. Phyloclim is a package implemented in R that integrates species ecological niche models with phylogenetics in order to calculate and visualize niche evolution on a phylogenetic tree. To do this, the user must first create a predicted species occupancy model. This involves using ecological niche modeling software, such as Maxent, and species location data stored as a raster file--I believe this is done in a mapping program like GIS. There is a great tutorial on the Maxent website.
Patterns of niche evolution or niche conservation can then be determined by comparing the variation between niche space within and between subclades. One of two outcomes is expected: 1) either niche evolution occurs between subclades with conservation of niche space within each subclade or 2) niche conservation occurs between subclades and niche evolution occurs within each subclade. This means you could either see a divergence of niche spaces at the beginning family followed by conservation of their niche spaces or you would see niche space preserved early on in evolutionary history followed by diversification among species. Inference can then be made about ancestral niche spaces using a statistical analysis, and a statement of how climate (or other variables) may or may not result in the diversification of a group.
I really liked using this R package. The visual of ancestral state reconstruction (see figure above) is really informative and neat to look at. However, Phyloclim is still quite new and no formal tutorial from the developers exists, but there are examples in the software package complete with example data. I have yet to try using my own data in Phyloclim, but I think it’s as simple reading a .csv file of your ecological niches models and your tree file in newick or nexus format (I like this tutorial) into R. I realize this may be easier said then done, but in theory it’s simple.
Jessica Craft
Useful papers:
Evans, M. E. K., S. A. Smith, R. S. Flynn, and M. J. Donoghue. 2009. Climate, niche evolution, anddiversification of the ’bird-cage evening primroses’ (Oenothera, sections Anogra and Kleinia). Am. Nat. 173: 225-240.
Fitzpatrick, B.M & Turelli, M. 2006. The geography of mammalian speciation:mixed signals from phylogenies and range maps. Evolution 60: 601-615.
Phillips, S.J, M. Dudik, & R.E. Schapire. 2006. Maximum entropy modeling of species geographic distributions. Ecological Modeling 190: 231-259.
Warren, D., R.E. Glor, & M. Turelli. 2008. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62: 2868-2883
 RSS Feed
RSS Feed
